A large family of anti‐activators accompanying XylS/AraC family regulatory proteins - Santiago - 2016 - Molecular Microbiology - Wiley Online Library
Functional Mechanism of the Efflux Pumps Transcription Regulators From Pseudomonas aeruginosa Based on 3D Structures

Characterisation of a putative AraC transcriptional regulator from Mycobacterium smegmatis - ScienceDirect

Transcriptional regulators: valuable targets for novel antibacterial strategies | Future Medicinal Chemistry

LysR family. (A) Architecture of the LTTR regulator and the regulated... | Download Scientific Diagram
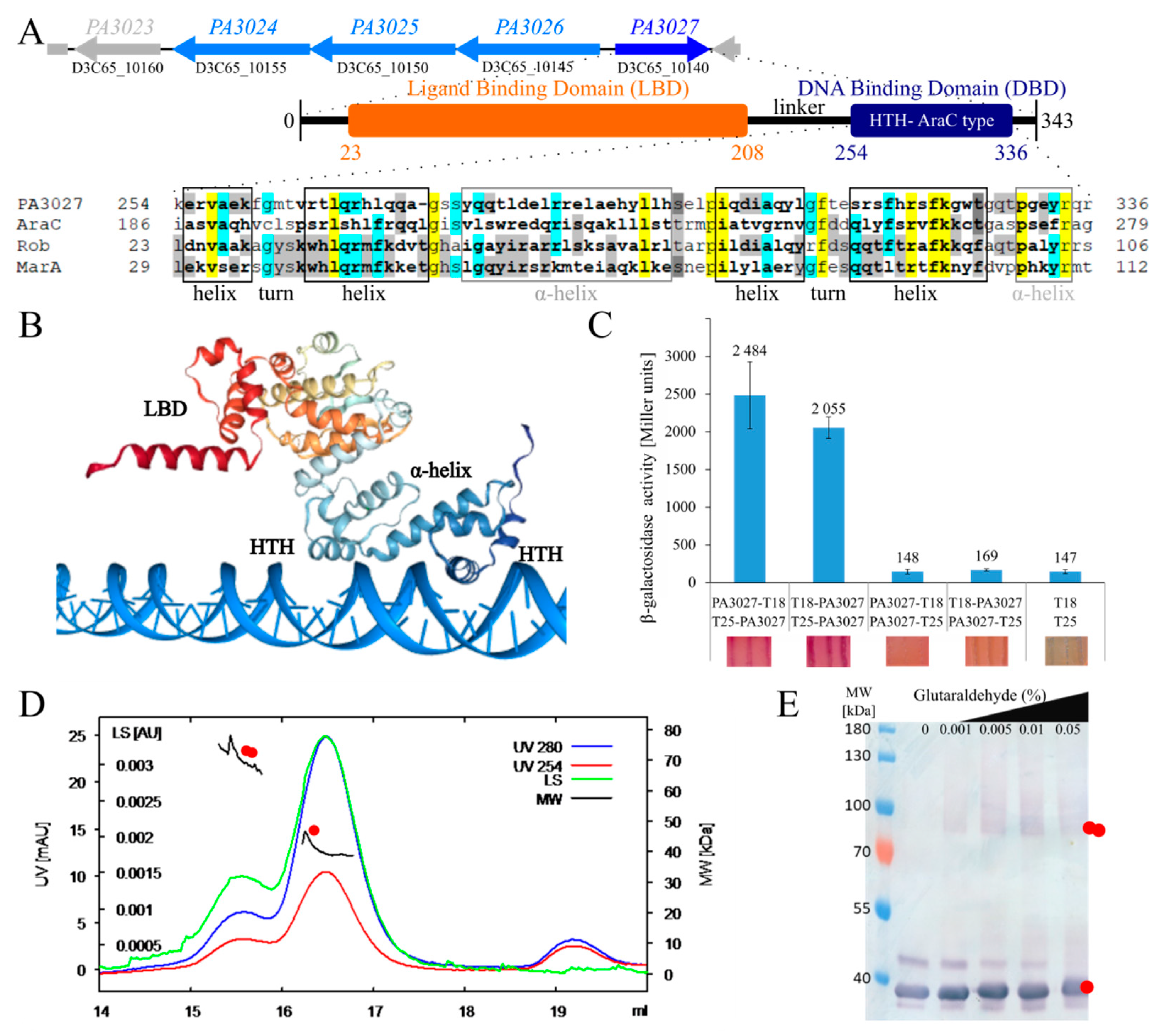
IJMS | Free Full-Text | The AraC-Type Transcriptional Regulator GliR (PA3027) Activates Genes of Glycerolipid Metabolism in Pseudomonas aeruginosa
![Escherichia coli transcription factors of unknown function: sequence features and possible evolutionary relationships [PeerJ] Escherichia coli transcription factors of unknown function: sequence features and possible evolutionary relationships [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2022/13772/1/fig-1-full.png)
Escherichia coli transcription factors of unknown function: sequence features and possible evolutionary relationships [PeerJ]
The AraC Negative Regulator family modulates the activity of histone-like proteins in pathogenic bacteria | PLOS Pathogens
The AraC Negative Regulator family modulates the activity of histone-like proteins in pathogenic bacteria | PLOS Pathogens

XylS-AraC family. (A) Activation mechanism. In the repression state,... | Download Scientific Diagram
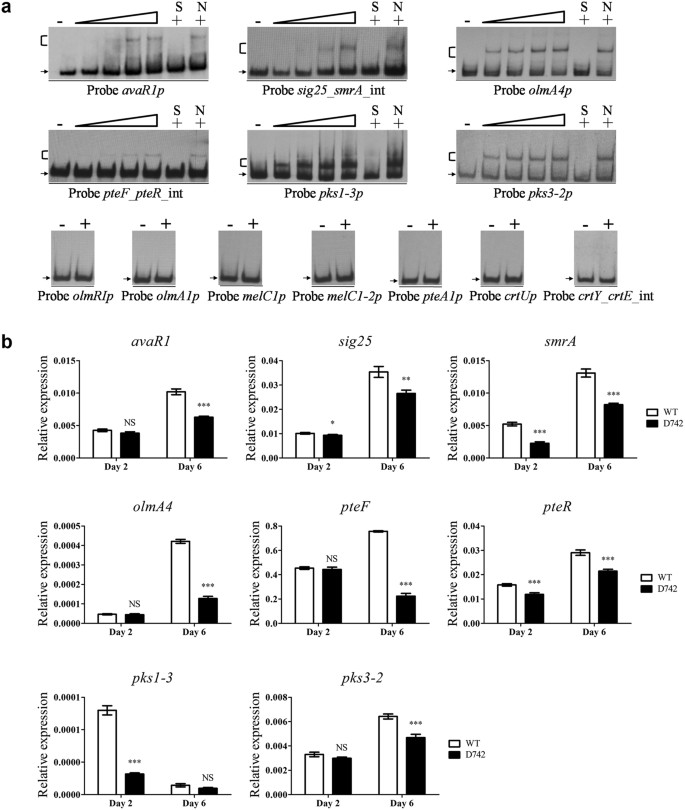
SAV742, a Novel AraC-Family Regulator from Streptomyces avermitilis, Controls Avermectin Biosynthesis, Cell Growth and Development | Scientific Reports

Modular, Multi-Input Transcriptional Logic Gating with Orthogonal LacI/GalR Family Chimeras | ACS Synthetic Biology

SAV742, a Novel AraC-Family Regulator from Streptomyces avermitilis, Controls Avermectin Biosynthesis, Cell Growth and Development | Scientific Reports

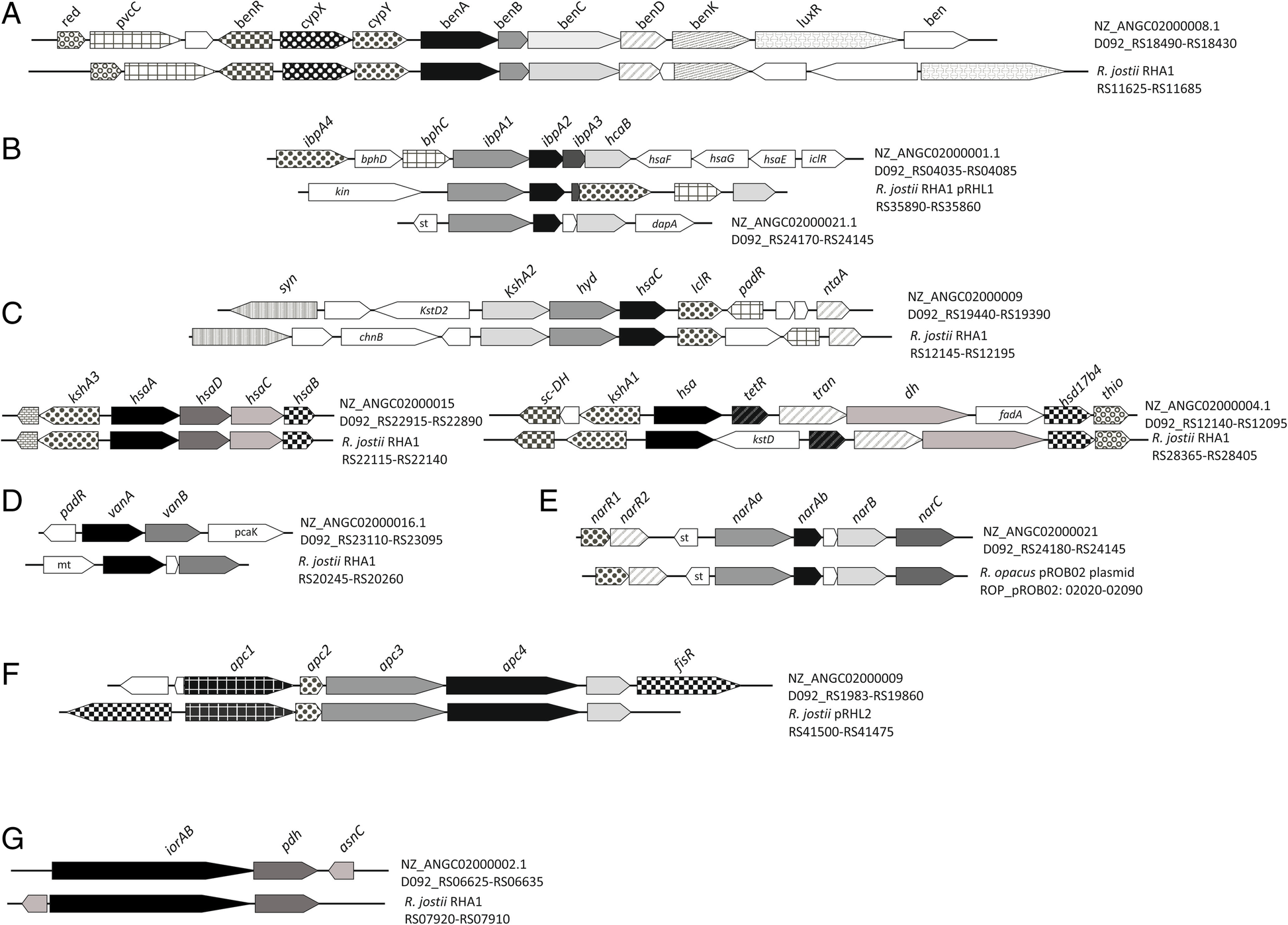
Mycobacterium tuberculosis Rv1395 Is a Class III Transcriptional Regulator of the AraC Family Involved in Cytochrome P450 Regulation* - Journal of Biological Chemistry

A large family of anti‐activators accompanying XylS/AraC family regulatory proteins - Santiago - 2016 - Molecular Microbiology - Wiley Online Library

AraC family of regulators. (A) Illustration of the "DNA-looping" and... | Download Scientific Diagram

Mechanisms used by AraC/XylS family regulators of viru- lence factor... | Download Scientific Diagram

An AraC/XylS Family Member at a High Level in a Hierarchy of Regulators for Phenol-Metabolizing Enzymes in Comamonas testosteroni R5 | Journal of Bacteriology